Increasing trends of antibiotic resistance, Uganda: an analysis of National antimicrobial resistance surveillance data, 20182021
Authors: Saudah Namubiru Kizito1*, Richard Migisha1, Paul Edward Okello1, Brenda Simbwa1, Zainah Kabami1, Brian Agaba1, Jane Frances Zalwango1, Helen Nelly Naiga1, Rebecca Akunzirwe1, Rita Namusosa2, Ibrahim Mugerwa2, Winnie Atuhaire2, Grace Najjuka2, Daniel Kadobera1, Alex Ario1, Susan Nabadda2; Institutional affiliations: 1Uganda Public Health Fellowship Program, National Institute of Public Health, Kampala, Uganda, 2National Health Laboratory and Diagnostic Services Department, Ministry of Health, Kampala, Uganda; *Correspondence: Email: skizito@musph.ac.ug, Tel: +256704518351
Summary
Background: Continuous monitoring of antimicrobial resistance (AMR) among isolates from clinical samples can inform effective drug selection for patients. In Uganda, human AMR surveillance occurs at national and regional referral hospitals and in selected public universities. Bacterial isolates from patients are subjected to drug susceptibility testing; the results are used in real-time for patient care. Isolates are then sent to the National Microbiology Reference Laboratory (NMRL) for re-analysis to generate national AMR surveillance data and for global reporting. Although isolate analysis results from NMRL are considered, the official AMR surveillance data, there is limited utilization of this data to inform public health planning. We evaluated trends and spatial distribution of AMR to common antibiotics used in Uganda.
Methods: We analyzed data from pathogenic bacterial isolates from blood, cerebrospinal, peritoneal, and pleural fluid from AMR surveillance data from 2018-2021. We calculated the proportions of isolates that were resistant to common antimicrobial classes. We use the chi-square test for trends to evaluate changes in AMR resistance over the study period.
Results: Out of 537 isolates with 15 pathogenic bacteria, 478 (89%) were from blood, 34 (6.3%) from pleural fluid, 21(4%) from cerebrospinal fluid, and 4 (0.7%) from peritoneal fluid. The most common pathogen was Staphylococcus aureus (20.1%), followed by Salmonella species (18.8%). Overall change in resistance over the four years was 63–84% for sulphonamides, fluoroquinolones macrolides (46–76%), phenicols (48–71%), penicillins (42–97%), β-lactamase inhibitors (20–92%), aminoglycosides (17–53%), cephalosporins (8.3–90%), carbapenems (5.3-26%), and glycopeptides- (0–20%). Annual resistance rates to ciprofloxacin increased from 2018-2021 for Gram-positive organisms (26-45% p=0.02). Among Gram-negative organisms, there were increases in resistance to tetracycline (29-78% p<0.001), ciprofloxacin (17-43%, p=0.004), ceftriaxone (8-72%, p=0.003), imipenem (6-26%, p=0.004), and meropenem (7-18, p=0.03).
Conclusions: There is a significant increase in the trends of drug resistance to antibiotics such as ciprofloxacin ceftriaxone, meropenem, imipenem, and tetracycline (among the Gram-negative organisms) in Uganda. Continuous monitoring of AMR trends at the national level to improve the efforts to reduce the AMR problem in Uganda through public health policy and planning is crucial.
Background
Antimicrobial resistance is the ability of microorganisms to resist the effects of medications that were previously used to treat such diseases[1]. It is a growing public health concern world over [2, 3]. Over 700,000 individuals worldwide die every year from illnesses linked to AMR. It is predicted that 10 million deaths will have occurred by 2050, costing the world $100 trillion [4].
Antimicrobial resistance has increased as a result of the use of broad-spectrum antibiotics to treat invasive infections, such as bloodstream and cerebrospinal fluid (CSF) infections. As a result, treating these infections is getting more challenging, which increases treatment failures and death [5].Over-the-counter medication access in underdeveloped countries like Uganda is one of the drivers of AMR in such nations[6]. Indiscriminate usage of antimicrobials exerts increased selection pressure on the bacterial population resulting in the accelerated emergence of AMR [7]. Antibiotic usage has led to the rise of germs that are widely multi-drug resistant, rendering even the most potent medications useless[8].
Recent estimates suggested that invasive infections, in particular antimicrobial-resistant invasive infections, account for 5.3 million deaths around the world annually. A significant proportion of these deaths occurs in low to middle-income countries (LMIC). While developing countries are battling an accelerated spread of AMR, developed countries are also experiencing the same trend [9].
The unavailability of reliable data in developing countries like Uganda makes it difficult to develop efficient methods to monitor and control AMR. Despite this urgent need to investigate AMR trends, only a handful of studies to date have reported [7] trends of resistance in Uganda. In a study that investigated antibiotic resistance in Uganda found that E. coli and K. pneumoniae was carried in the gut of clients attending outpatient clinics in Kampala and two rural districts [10]. It further showed high rates of resistance to commonly used antibiotics such as ampicillin and septrin and relatively lower resistance rates to amoxicillin/ clavulanate, chloramphenicol, ciprofloxacin, gentamicin, nitrofurantoin and ceftriaxone.
We described the organisms that are isolated from patients’ sterile site samples and their antibiotic resistance trends in Uganda, to inform planning and AMR control interventions.
Methods
Study setting, design, and data source
We conducted a descriptive analysis of Uganda’s national AMR surveillance data, 2018-2021. National surveillance sites for AMR in human health comprise of microbiology laboratories of Regional referral hospitals, National referral hospitals, and selected institutions of learning. These sites analyze clinical samples (pathogen identification and Antimicrobial susceptibility testing -AST) for routine patient care. The decision to do a bacterial culture test is entirely at the discretion of the attending clinician. For quality control purposes, these sites also refer isolates with their relevant identifiers to the National Microbiology Reference Laboratory (NMRL) (at the National Health Laboratory and Diagnostic Services (NHLDS) department of the Ministry of Health) for re-identification and antimicrobial susceptibility re-testing. NMRL is accredited by the College of American Pathologists (CAP) in accordance with the recognized international standard ISO 15189:2012. The accredited microbiology tests include microscopy, culture, identification, and antibiotic sensitivity tests. The laboratory’s main catchment area is national.
Inclusion and exclusion
All Isolates received from all the surveillance sites as patient samples were re-analyzed by the laboratory. However only isolates with re-identification results matching with primary sites were analyzed in this study.
Study variables
We extracted individual-level data from the NMRL database. Data was extracted as an excel sheet from this electronic database containing patients’ reports, organism identified, and sentinel sites. Information obtained on each case included: patient’s age, patient’s sex, year of sample collection, specimen source, hospital in which the sample was drawn, isolated pathogen in the positive culture, and susceptibility results (defined as susceptible, intermediate, or resistant).
Data management and analysis
The variables analyzed included the age, sex, specimen type (i.e., blood, and cerebrospinal fluid), organisms isolated, and antibiotics tested (i.e., ampicillin, amoxicillin, augmentin, ciprofloxacin, cotrimoxazole, ceftriaxone, penicillin, gentamicin, carbapenems, vancomycin). The outcome was the antibiotic resistance of the isolates to common antibiotics. Isolates were classified as either susceptible or resistant to an antimicrobial and all isolates with intermediate reactions were classified as resistant. Data quality was checked using the completeness of data entries in the Laboratory Information System. Rates of resistance were calculated as a proportion of resistant organisms out of total number of that organisms that is tested for antimicrobial susceptibility per year. Resistance to each antibiotic was analyzed separately and Microsoft Excel 2016 was used to plot the trends from 2018 to 2021. Chi square test for trends was used to test the significance of antibiotic resistance trends over time. Epi-Info™ 7- (CDC, Atlanta, Georgia) statistical package was used for additional statistical analysis. The p-value < 0.05 was considered statistically significant.
Ethical considerations
We obtained permission to use data from the NHLDS. Data was stored on a password protected computer and only accessed by the study team. Data abstracted did not have individual unique identifier information. We also obtained a non-research determination from the US CDC
Results
Attributes of organisms isolated from sterile sample cultures, Uganda, 2018-2022
Of the 537 isolates analyzed, 188(52.6%), were from children were 0-5years followed by 67(18.7%) which were from persons 6-18years then 19-45years 59(16.2%) and were from adults above 66years. Fifty-five percent of isolates were from females. Over time, 2018 had the highest number of isolates 151(28.1%), while 2019 had the least number of isolates 93(17%). Geographically, most isolates were from Mbarara Regional Referral Hospital 186(48%) while the least came from Kiruddu National Referral Hospital and the General Military Hospital, each having 1(0.3%). 108(20.1%) of isolates were Staphylococcus aureus followed by Salmonella species which were 101(18.8%) (Table 1).
Table 1: Attributes of organisms isolated from sterile sample cultures, Uganda, 2018-2022
| Variable | Number of pathogens (n=537) | Percent |
| Age(years) *n=357 | ||
| 0-5 | 188 | 52.7 |
| 6-18 | 67 | 18.8 |
| 19-45 | 59 | 16.5 |
| 46-65 | 26 | 7.3 |
| 66 and above | 17 | 4.8 |
| Sex *n=480 | ||
| Male | 216 | 45 |
| Female | 264 | 55 |
| Sample type analyzed | ||
| Blood | 478 | 89 |
| Cerebral spinal fluid | 21 | 3.9 |
| Peritoneal fluid | 4 | 0.7 |
| Pleural fluid | 34 | 6.3 |
| Year of isolation | ||
| 2018 | 151 | 28.1 |
| 2019 | 93 | 17.3 |
| 2020 | 179 | 33.3 |
| 2021 | 114 | 21.2 |
| Location of primary testing laboratory | ||
| Arua RRH | 35 | 6.5 |
| Fortportal RRH | 50 | 9.3 |
| Hoima RRH | 6 | 1.1 |
| General Military Hospital Bombo | 1 | 0.2 |
| 1DI/DMM-Makerere | 139 | 25.9 |
| Jinja RRH | 38 | 7.1 |
| Kabale RRH | 24 | 4.5 |
| Kiruddu N | 1 | 0.2 |
| Mbale RRH | 17 | 3.2 |
| Mbarara RRH | 217 | 40.4 |
| Soroti RRH | 8 | 1.5 |
| Gram class of organisms | ||
| Negative | 351 | 65 |
| Positive | 186 | 35 |
| Organisms isolated *n=534 | ||
| A Cinetobacter spp | 22 | 4.1 |
| Citrobacter spp | 11 | 2.1 |
| Enterobacter spp | 25 | 4.7 |
| Enterococcus spp | 59 | 11.1 |
| Escherichia coli | 80 | 15 |
| Klebsiella pneumoniae | 83 | 15.5 |
| Proteus spp | 9 | 1.7 |
| Pseudomonas aeruginosa | 15 | 2.8 |
| Salmonella typhii/paratyphii | 101 | 18.9 |
| Staphylococcus aureus | 108 | 20.1 |
| Streptococcus spp | 14 | 2.6 |
| Others | 7 | 1.3 |
1DI/DMM=Infectious Diseases Institute/Department of Medical Microbiology
RRH=Regional Referral Hospital
NRH=National Referral Hospital
Spp = species
Others=Bacillus spp, Morganella morganii, Providencia spp
Trends of total antimicrobial resistance of Gram-positive organisms to common antibiotics, Uganda, 2018–2021
Increasing resistance to Ciprofloxacin over the four-year study period was observed (26.2-44.9%; p < 0.01) (Figure 1).
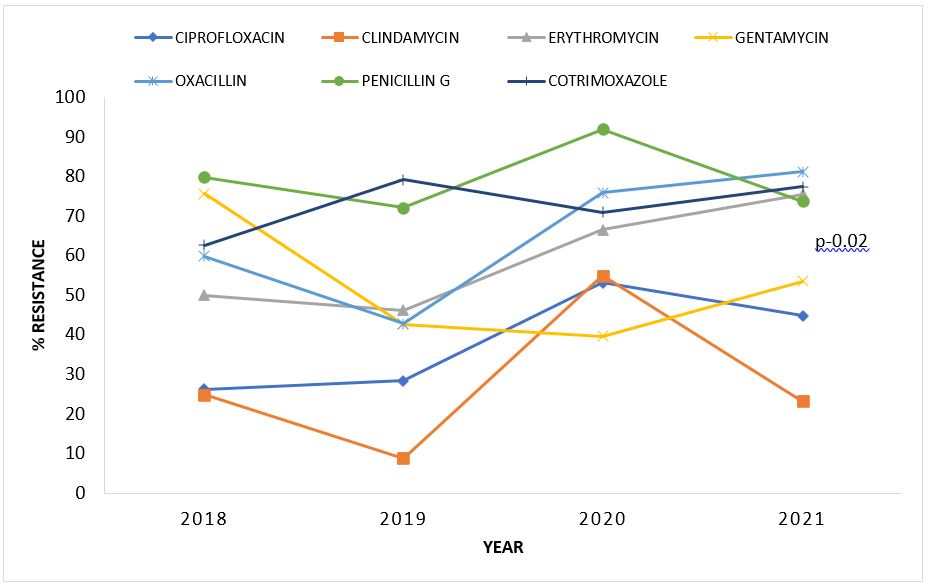
Rates of resistance of Gram-negative organisms to common antibiotics, Uganda 2018–2021
Overall, percentage antibiotic resistance of Gram-negative organisms increased over the years. Resistance to penicillins (Piperacillin and Ampicillin) was highest among Gram negative bacteria. Specifically, increase in resistance over years were noted for Ceftriaxone (8.3-78.6%; p-0.003), Ciprofloxacin (17.1-42.6%; p-0.004), Imipenem (5.7-29.7%; p-0.004), Meropenem (5.3-18.4%; p-0.03), and Tetracycline (28.6%-81.6%; p-0.02) (Table 2).
Table 2: Rates of resistance of Gram-negative organisms to common antibiotics, Uganda, 2018-2021
| Antibiotic/Year | 2018
n (%) |
2019
n (%) |
2020
n (%) |
2021
n (%) |
Chi2 Value for Trends | p-value |
| Amikacin | 11(52.4) | 8(22.9) | 12(20.3) | 14(38.9) | 0.24 | 0.63 |
| Ampicillin | 41(62.1) | 35(89.7) | 58(89.2) | 19(76) | 0.93 | 0.34 |
| Amoxiclav | 6(46.2) | 23(54.8) | 28(87.5) | 23(92) | 2.83 | 0.09 |
| Cefotaxime | 9(39.1) | 20(60.6) | 44(78.6) | 34(87.2) | 3.39 | 0.07 |
| Ceftriaxone | 3(8.3) | 25(62.5) | 66(78.6) | 41(71.9) | 8.77 | <0.001 |
| Cefoxitin | 4(80) | 8(25.4) | 14(33.3) | 4(50) | 0.01 | 0.92 |
| Cefepime | 13(56.5) | 13(50) | 48(71.6) | 29(90.6) | 1.95 | 0.16 |
| Chloramphenicol | 28(48.3) | 26(51) | 51(71.6) | 26(48.1) | 0.03 | 0.87 |
| Ciprofloxacin | 12(17.1) | 16(30.2) | 48(5.8) | 20(42.6) | 8.35 | <0.001 |
| Gentamycin | 15(75) | 16(45.7) | 21(45.7) | 14(70) | 0.04 | 0.84 |
| Imipenem | 4(5.7) | 3(8.8) | 11(19) | 14(25.9) | 8.42 | <0.001 |
| Meropenem | 5(7.2) | 2(5.3) | 7(18.4) | 5(2.5) | 4.82 | 0.03 |
| Piperacillin | 20(76.9) | 38(97.4) | 36(87.8) | 0.07 | 0.79 | |
| TPZ | 1(20) | 4(57.1) | 13(68.4) | 15(68.2) | 0.80 | 0.37 |
| SXT | 60(62.5) | 37(84.1) | 59(78.7) | 31(75.6) | 0.68 | 0.41 |
| Tetracycline | 14(28.6) | 23(71.6) | 18(81.8) | 14(77.8) | 0.5 | 0.02 |
TPZ= Piperacillin tazobactam
SXT=Sulfamethoxazole trimethoprim
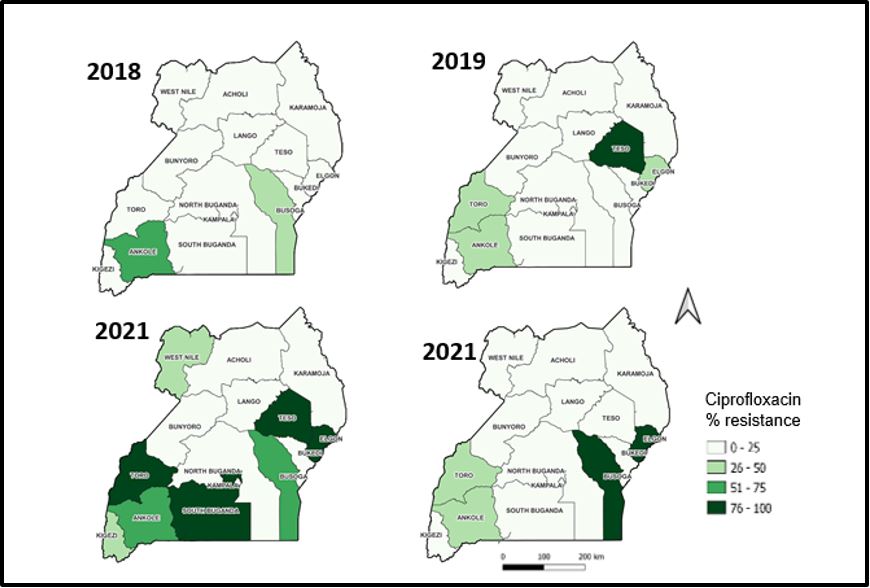
Trends of resistance to ciprofloxacin by health region, 2018-2021
There was increasing resistance to Ciprofloxacin across the health sub-regions. The significant decline observed in 2021.There is an increasing resistance that is observed in the Elgon sub-region over time, starting with pale green in 2018, and increasing to dark green in 2020 which is sustained in 2021 (Figure 2).
Discussion
We reported trends and patterns of antimicrobial resistance of organisms isolated from patients’ sterile site samples in Uganda, 2018-2021 with an emphasis on the antibiotics frequently utilized to treat common infections and the bacterial Gram classification. We found increases in rates of resistance to commonly used antibiotics over time. The most commonly encountered Gram-positive organisms was Staphylococcus aureus while the commonest Gram-negative organisms were Salmonella typhii/paratyphii species and Escherichia coli.
Gram positive organisms were increasingly resistant to Ciprofloxacin and Gram-negative organisms increasing resistance to Ceftriaxone, Ciprofloxacin, and Tetracycline as well as last resort antibiotics like Meropenem and Imipenem. The growing resistance rates to carbapenems is worrisome and may lead to the spread of fatal infections especially in hospitals[11].
Although a number of studies have previously detailed the susceptibility rates in a number of infectious isolates, Ugandan data are restricted to either single-center studies or to research focusing on the susceptibility to specific patient categories, wards, organism groups, or antibiotic classes. There is limited utilization of nationwide AMR surveillance data. This is important because lack of nationwide trends of resistance, and underutilization of routine surveillance data may slow AMR control due to the lack of generalizable data.
The increasing in resistance to these antibiotic classes over time is consistent with literature which states that the more an antibiotic is used, the more the bacteria can develop resistance against it[12].These findings are similar to those obtained from the Global Point Prevalence Survey on antimicrobial use and stewardship study carried out in 2021, where 52% of prescriptions in Uganda were from WHO Access class of antibiotics while 39% were from the Watch class of antibiotics[13]. Also, in a study in Tertiary care facilities in Uganda, 50.6% of participants were prescribed Ceftriaxone while 25.5% were prescribed Levofloxacin [14].
Our study showed that over the course of four years, both Gram-positive and Gram-negative bacteria had an increase in resistance to ciprofloxacin from 26 to 56 percent. This is similar to the findings from a systematic review that included studies addressing the prevalence of ciprofloxacin-resistant clinical isolates in Ethiopia which revealed that one in five clinical isolates were found to be ciprofloxacin resistant in both gram-positive and gram-negative bacteria [15]. A similar study done in Ethiopia, in 2015 observed that the rates of resistance to Ciprofloxacin were high (20-70%) among bacterial organisms isolated from blood which is a sterile site sample[16].
Another salient finding is the increase in resistance of Gram-negative organisms to antibiotics; Ceftriaxone (8-79%), Tetracycline (28-81%), and Imipenem (6-26%). This is similar to other Ugandan/regional resistance rates. Ampaire et al conducted a systematic review in 2018 where he found that there is an overall increase in resistance to Cefotaxime (46-49%) and Cefuroxime both of which are third generation Cephalosporins, whose resistance can be used to infer Ceftriaxone resistance [17]. On the contrally, in this same study Ciprofloxacin was found to be generally susceptible and hence the recommended drug of choice for infections caused by Gram negative organisms. Mackay and others, who conducted a 12-month AMR survey from October 2011 to September 2012 at a tertiary facility in Cape Town, South Africa, [18] found that for health-care associated Enterobacteriaceae bloodstream isolates, susceptibility rates were 58.5% to ceftriaxone, and 70% to ciprofloxacin.
Study limitations
We only analyzed data for pathogenic bacterial isolates from blood, cerebrospinal fluid peritoneal fluid, and pleural fluid. Other sources of bacterial infections such as urinary tract infections were not included in this analysis.
Conclusion
There is a significant increase in the trends of drug resistance to antibiotics such as ciprofloxacin ceftriaxone, meropenem, imipenem, and tetracycline (among the Gram-negative organisms) in Uganda. Continuous monitoring of AMR trends at the national level to improve the efforts to reduce the AMR problem in Uganda through public health policy and planning is crucial.
Conflict of Interest
The authors declare that they had no conflict of interest.
Acknowledgments
The authors would like to thank Ministry of Health who initiated questions that led to this analysis, Department of Laboratory and Diagnostic Services, National Microbiology Reference Laboratory and all regional and national referral hospital microbiology laboratories who generated the data that we analyzed.
Copyrights
All materials in this manuscript is in the public domain and may be used and reprinted without permission; citation as to source; however, is appreciated. Any article can be reprinted or published. If cited as a reprint, it should be referenced in the original form.
References
- Uddin, T.M., et al., Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. Journal of infection and public health, 2021. 14(12): p. 1750-1766.
- !!! INVALID CITATION !!! [1].
- Sugden, R., R. Kelly, and S. Davies, Combatting antimicrobial resistance globally. Nature microbiology, 2016. 1(10): p. 1-2.
- Mhondoro, M., et al., Trends in antimicrobial resistance of bacterial pathogens in Harare, Zimbabwe, 2012–2017: a secondary dataset analysis. BMC Infectious Diseases, 2019. 19(1): p. 746.
- Javaid, N., et al., Trends in antimicrobial resistance amongst pathogens isolated from blood and cerebrospinal fluid cultures in Pakistan (2011-2015): A retrospective cross-sectional study. PLOS ONE, 2021. 16(4): p. e0250226.
- Ayukekbong, J.A., M. Ntemgwa, and A.N. Atabe, The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrobial Resistance & Infection Control, 2017. 6(1): p. 1-8.
- Ferri, M., et al., Antimicrobial resistance: A global emerging threat to public health systems. Critical reviews in food science and nutrition, 2017. 57(13): p. 2857-2876.
- Santajit, S. and N. Indrawattana, Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res Int, 2016. 2016: p. 2475067.
- !!! INVALID CITATION !!! [5].
- Najjuka, C.F., et al., Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC research notes, 2016. 9: p. 1-14.
- Nordmann, P., T. Naas, and L. Poirel, Global spread of carbapenemase-producing Enterobacteriaceae. Emerging infectious diseases, 2011. 17(10): p. 1791.
- Llor, C. and L. Bjerrum, Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Therapeutic advances in drug safety, 2014. 5(6): p. 229-241.
- D’Arcy, N., et al., Antibiotic prescribing patterns in Ghana, Uganda, Zambia and Tanzania hospitals: results from the global point prevalence survey (G-PPS) on antimicrobial use and stewardship interventions implemented. Antibiotics, 2021. 10(9): p. 1122.
- Kizito, M., et al., Antibiotic Prevalence Study and Factors Influencing Prescription of WHO Watch Category Antibiotic Ceftriaxone in a Tertiary Care Private Not for Profit Hospital in Uganda. Antibiotics, 2021. 10(10): p. 1167.
- Sisay, M., et al., Resistance profile of clinically relevant bacterial isolates against fluoroquinolone in Ethiopia: a systematic review and meta-analysis. BMC Pharmacology and Toxicology, 2018. 19: p. 1-14.
- Ulhas, A.A., Prevalence and Characterization of carbapenem resistant organisms causing urinary tract infections among hospitalized patients and outcomes of these infections in a tertiary care center. 2018, Christian Medical College, Vellore.
- Ampaire, L., E. Nduhura, and I. Wewedru, Phenotypic prevalence of extended spectrum beta-lactamases among enterobacteriaceae isolated at Mulago National Referral Hospital: Uganda. BMC research notes, 2017. 10(1): p. 1-4.
- Carroll, M., et al., Five-year antimicrobial susceptibility trends among bacterial isolates from a tertiary health-care facility in Kigali, Rwanda. The American journal of tropical medicine and hygiene, 2016. 95(6): p. 1277.

